Spot the difference: genomic sub-types of leukaemia
Recent genetic analysis exposes differences in blood cancer, offering hope for patients and the future of personalised medicine
Cancer is, essentially, a genetic disease – caused by the accumulation of mutations in a cell’s DNA that allow it to evade the normal controls over how, when and where it can multiply and specialise. The advent of affordable genome sequencing means that oncology and haematology are rapidly evolving as our understanding of cancer grows; the ultimate goal is better treatments, though tools to improve risk prediction or provide earlier and more precise diagnosis and prognosis are also highly valuable.
The fight against blood cancer
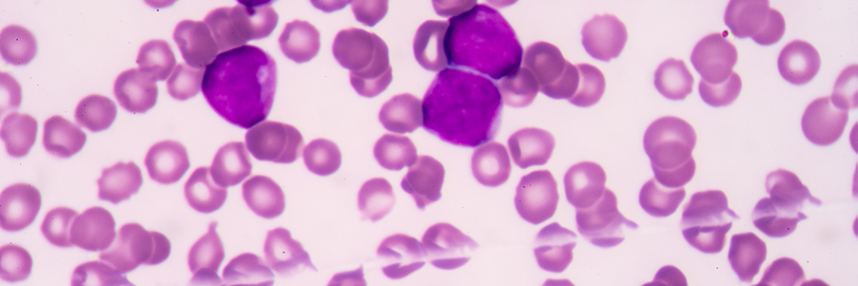
New research led by the Sanger Institute may help the fight against acute myeloid leukaemia (AML), a rare and aggressive form of blood cancer caused by mutations in the bone marrow stem cells that give rise to myeloid white blood cells. The normal function of these cells is in immune defence; AML patients have sharply reduced resistance to bacterial and parasitic infections. Overall long-term survival for the cancer is quite poor, but outcomes vary sharply between different patients; those aged over 65, with recurrent disease or AML that arises following radiotherapy for a previous cancer tend to do less well.
Previous work has revealed that cancer cells from AML patients can contain a huge number and variety of different mutations. This made it more difficult to identify which play an important role in cancer progression (‘driver’ mutations) and which are less significant (‘passenger’ mutations), but efforts such as the US Cancer Genome Atlas have created a large database of known AML mutations. Historically, AML cases were classified on the basis of the physical features (morphology) of the cancer cells, but it has become apparent that it makes more sense to group cases that share common genetic origins together as sub-groups of cancer.
Untangling the origins of AML
For the new study, scientists examined over 100 AML mutations from more than 1500 patients to search for shared genomic profiles, or signatures. They found at least eleven distinct groups of related genomic features, leading them to conclude that what has previously been classed as one disease is effectively at least eleven distinct (though related) forms of AML. Identifying which of these genomic sub-groups a case of AML belongs to allows a modest improvement (about 7%) in the ability to predict the likely course of disease. However, it seems that the precise set of mutations present in an individual patient’s AML cells, whilst falling into one of eleven broad groupings, may be unique – which may explain the very variable outcomes seen in different patients.
Co-author of the research published in the New England Journal of Medicine Dr Elli Papaemmanuil said that they had “untangled the genetic complexity seen in most AML cancer genomes into distinct evolutionary paths” that lead to the final cancer.
Ideally, this information will be useful for segregating AML patients with shared genomic profiles for clinical trials of potential new treatments. The researchers will need to test their findings by analysis of DNA from a fresh group of AML patients, to check whether they are reliable, but if so, they hope that genomic analysis and classification will become widely available to patients as plans to integrate genomics across the NHS in England advance.
–