Microarray (array CGH)
Microarray is a high-resolution genome-wide screen for copy number variants.
Introduction
Microarray is a method that tests for copy number variation (deletions and duplications) across the genome. It is considered the first-line investigation for patients with developmental delay, intellectual disability, autism, epilepsy or congenital abnormalities. It can also be used in the prenatal setting for the investigation of fetal ultrasound anomalies.
How does microarray work?
DNA microarrays detect copy number alterations across the genome. There are two different ways of performing microarray testing: array comparative genomic hybridisation (aCGH) and single nucleotide polymorphism array (SNP array).
aCGH
- Patient and reference DNA are labelled with different coloured fluorescent dyes (usually red for reference and green for patient DNA).
- An array slide is spotted with oligonucleotide DNA probes – small molecules of DNA that are designed to hybridise with a particular section of the genome. Probe distribution around the genome is not even. The probes provide ‘backbone’ coverage around the genome, and additional probes may be present in gene rich regions, syndrome regions or specific genes of interest.
- Patient and reference DNA are washed over the array slide and bind competitively to the probes.
- The array slide is washed and scanned, and the fluorescence emitted at each probe location is measured.
- Computer software then analyses the data:
- Yellow: equal patient and reference DNA present.
- Red: more reference DNA than patient DNA, therefore a deletion is present.
- Green: more patient DNA than reference DNA, therefore a duplication is present.
- This process is summarised in figure 1, below.
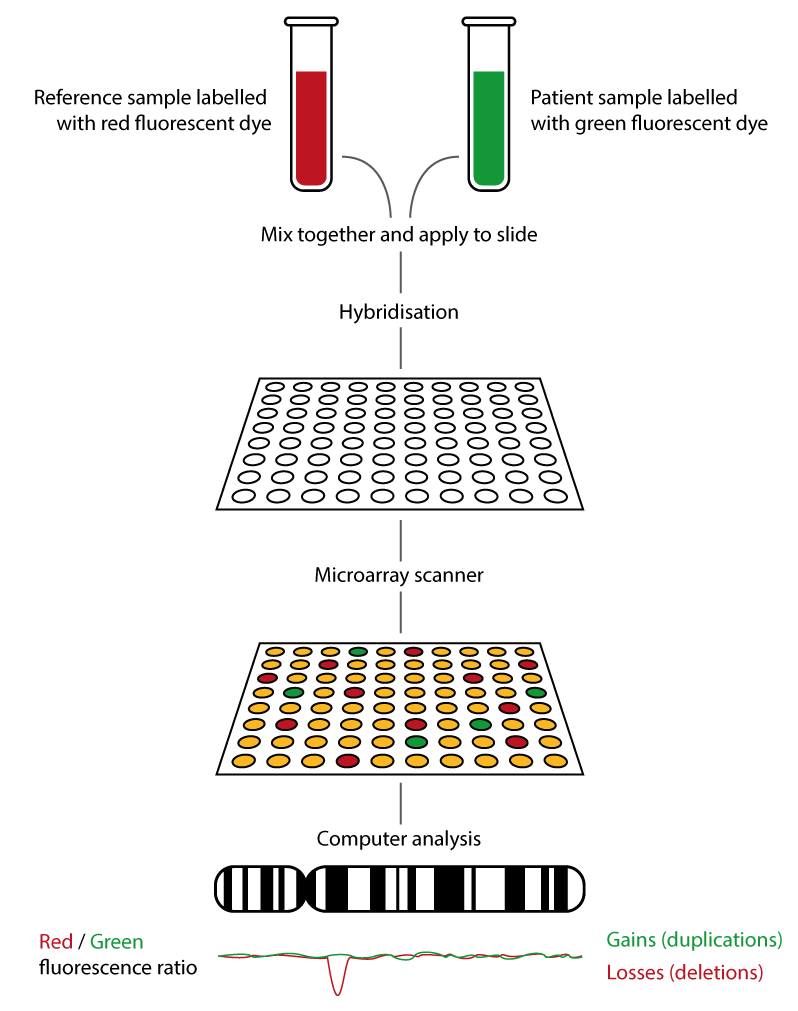
Figure 1: How microarray (array CGH) works
SNP array
- Single nucleotide polymorphisms (SNPs) are common genetic differences between individuals.
- A SNP array slide is spotted with allele-specific DNA probes targeting regions in which there is SNP variation between individuals.
- Patient DNA is hybridised to the array slide and binds to the probes.
- The array slide is scanned, and the fluorescence emitted at each probe location is measured.
- The fluorescence emitted is dependent on which alleles are present in the patient at the SNP site targeted by the probe.
- Computer software then analyses the data, identifying regions in which copy number variation is present by looking at which nucleotides are present at each SNP location.
- Individuals may be heterozygous (AB or BA) or homozygous (AA or BB) for each SNP. Where a deletion is present, every SNP in the deleted region will appear homozygous and there will be a loss of signal intensity. Where a duplication is present, the ratio of alleles in the duplicated region will be different to that in normal regions, and there will be a gain of signal intensity.
Hybrid arrays
SNPs do not occur evenly across the genome. This can lead to SNP arrays with gaps in coverage. Therefore, combined SNP and oligonucleotide arrays are sometimes used. In this method, oligonucleotide probes are used alongside probes targeting SNPs, which evens out coverage and can allow targeting of specific syndrome-associated regions or genes.
Advantages and limitations of microarray
Advantages
- High resolution copy number variants (approximately 50–200Kb, depending on coverage) can be detected; this is much higher than karyotyping, in which resolution is around 5Mb
- Accuracy of copy number variant detection is higher than in next-generation sequencing-based assays, including whole genome sequencing (WGS).
- Genome-wide test: no need to target a specific imbalance, in contrast to other copy number variant detection methods such as FISH or MLPA.
- Simultaneous processing of large numbers of samples.
- SNP arrays only: can have higher resolution than aCGH; can detect copy number neutral loss of heterozygosity, some forms of uniparental disomy, and ploidy changes.
Limitations
- Microarray cannot detect balanced translocations or inversions, so karyotyping is used for these, for example in the investigation of infertility or recurrent miscarriage.
- Microarray cannot provide structural or positional information. For example, where a duplication is detected, it will not be clear from the microarray whether this is in tandem (next to the original copy) or elsewhere in the genome. Karyotyping or FISH may be used to provide positional information.
- Small copy number changes such as exon deletions and duplications within a gene may not be detected. An alternative method such as MLPA may be more appropriate if the gene to be investigated is known.
- Microarray is not a sequencing test. It will not pick up sequence changes in a gene. Whole genome sequencing has the potential to detect both copy number variation and sequence variation in a single assay.
- Mosaicism may not be reliably detected.
- In prenatal diagnosis, more expensive and slower than common aneuploidy testing by QF-PCR (therefore QF-PCR is usually used as a first line test, with microarray used if QF-PCR is negative in the presence of ultrasound anomalies).
- aCGH specific limitations: cannot detect triploidy, uniparental disomy or copy-number neutral loss of heterozygosity.
- SNP array specific limitations: coverage is limited to regions where there is SNP variation between individuals.
Key messages
- Microarray is a high resolution test for copy number variants.
- Sequence changes will not be detected because microarray is not a sequencing test.
- Microarray is the first line investigation for patients with developmental delay, intellectual disability, autism, epilepsy or congenital abnormalities.
- There are two types of microarray: array comparative genomic hybridisation (aCGH) and single nucleotide polymorphism array (SNP array).
Resources
For clinicians
- Science by Nature: Microarray-based Comparative Genomic Hybridization (aCGH)