Karyotype
A karyotype is a visual representation of the number and structure of all the chromosomes and provides a low-resolution genome-wide screen for chromosomal variants.
Overview
A karyotype provides a visual, genome-wide screen for chromosomal variants such as deletions, duplications and structural rearrangements. It has a limited resolution of 5–10 megabases (Mb), which means that it cannot detect variants that are smaller than this.
In most contexts, karyotyping has been superseded by the use of microarray, which can also detect chromosomal variants but at much higher resolution (50–200 kilobases (Kb)). Nevertheless, karyotyping is still in use in certain clinical situations.
Clinical applications
One such clinical situation is in the investigation of infertility or recurrent miscarriage. Here, karyotyping has an advantage over microarray because it is able to detect balanced rearrangements, which can lead to infertility. Karyotyping can do this because it provides an image of the structure of whole chromosomes, unlike microarray, which shows the amount of chromosomal material present but not its position.
Another common application of karyotyping is the detection of structural chromosome rearrangements resulting in gene fusions that drive cancer. An example is the Philadelphia chromosome in chronic myeloid leukaemia (a translocation of chromosomes 9 and 22).
What genomic variants can a karyotype identify?
Aneuploidy (numerical variants in the chromosomes, where a chromosome is missing or is present in one or more extra copies) can be detected by karyotyping. Examples of this include:
- trisomy of chromosomes 13, 18 or 21; and
- sex chromosome variants, such as Turner syndrome (45,X) or Klinefelter syndrome (47,XXY).
Karyotyping can also detect structural rearrangements (with a limit of resolution of 5Mb), including:
- translocations;
- inversions;
- deletions (larger than 5Mb);
- duplications (larger than 5Mb);
- ring chromosomes; and
- marker chromosomes (small, structurally abnormal, unidentifiable chromosomes).
A clinical karyotype report will include a description of the cytogenetic findings, described using standardised nomenclature and displayed using a cartoon representation of the chromosome and its banding pattern, called an ideogram (see figure 1).
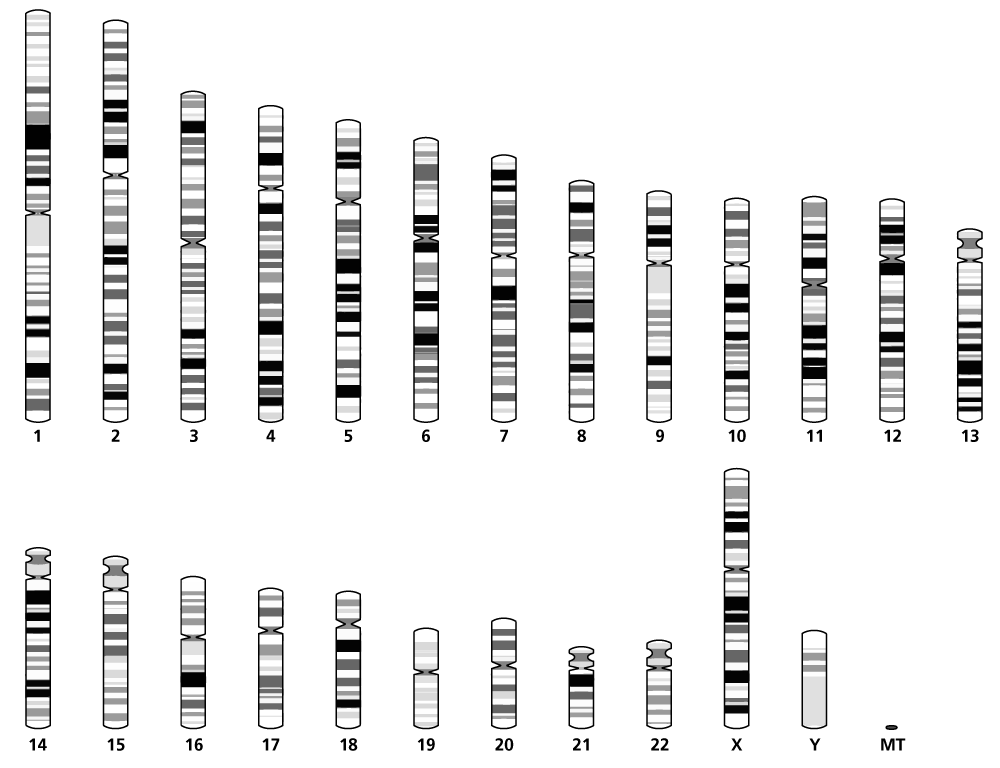
Figure 1: An ideogram showing one of each chromosome. Image adapted from Ensembl. Cunningham F, Allen JE, Alvarez-Jarreta J and others. ‘Ensembl 2022′. Nucleic Acids Research 2022: volume 50, issue D1, pages D988–D995. DOI: 10.1093/nar/gkab1049
How does it work?
Karyotyping can be done on any sample from which nucleated cells (which can be cell cultured – that is, grown in a laboratory) can be obtained, most commonly:
- blood;
- skin;
- prenatal samples (such as chorionic villus or amniotic fluid); and
- bone marrow.
Cell culturing is required for karyotyping. This can take anything from three days (blood and bone marrow) up to 7 to 14 days (skin and prenatal samples). The process is outlined below.
- Cells are grown in an environment that includes stimulants for cell division (this may not be required for cancer cells, which divide rapidly themselves).
- Cell growth is arrested at metaphase and harvested for analysis.
- Once harvested, the cells are mounted on slides, treated with enzymes and Giemsa stain to produce G-banding patterns, then viewed under a light microscope (x1,000).
- The banding patterns for each different chromosome are characteristic and reproducible, resembling a barcode.
- The cytogeneticist identifies and counts the chromosomes, ‘cuts and pastes’ the image to pair the homologues, and compares the banding pattern in each pair to identify any rearrangements, deletions or duplications.
Many attempts have been made to develop computer software to replace this labour-intensive work, but it has not been possible to replace the pattern-recognition skills of a highly trained cytogeneticist.
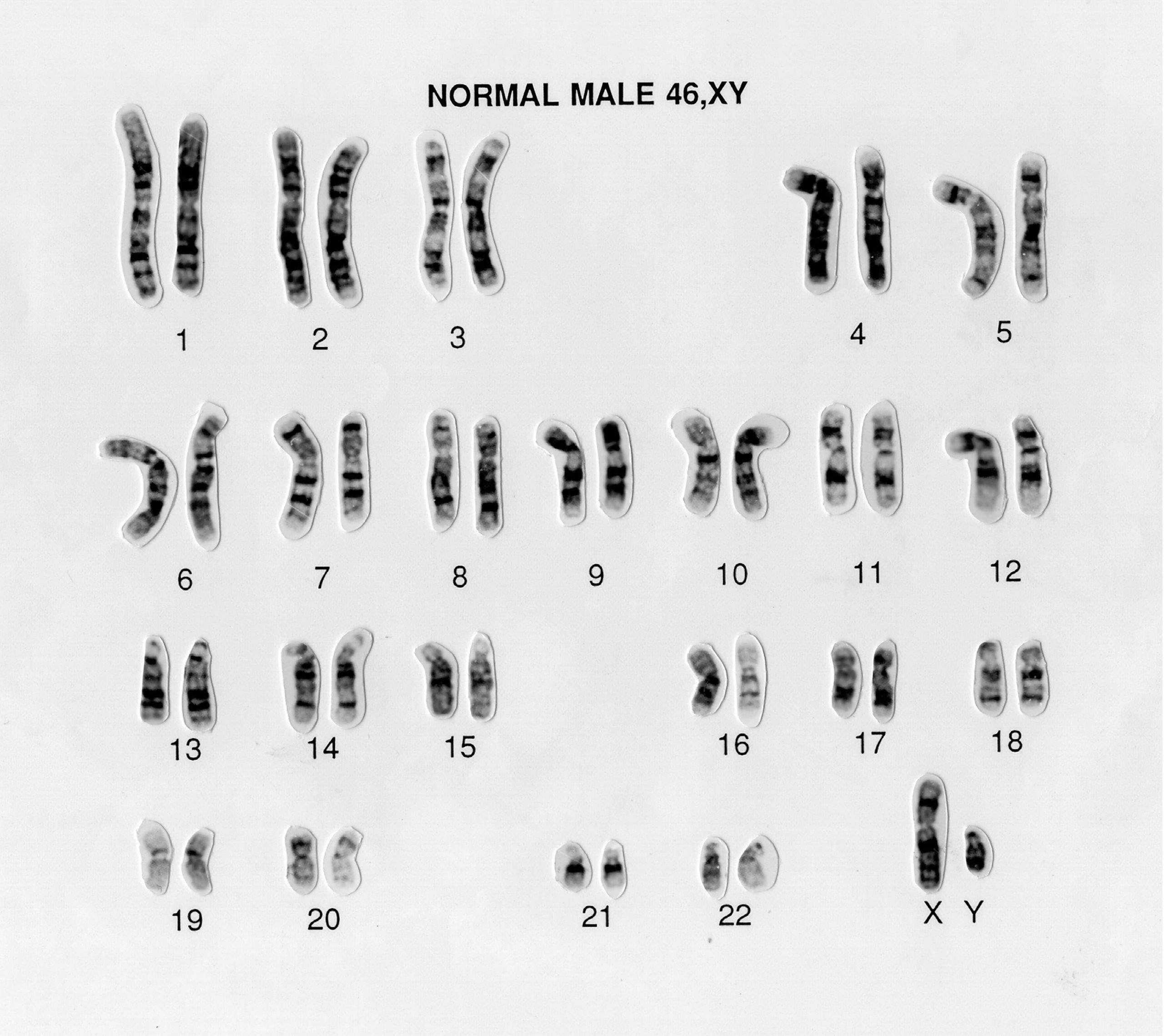
Figure 2: A normal male karyotype (note the X and Y sex chromosomes). Source: Wellcome Collection (CC BY 4.0).
Advantages and limitations of karyotyping
Advantages
Karyotyping:
- screens the whole genome;
- detects balanced and unbalanced rearrangements;
- provides positional information;
- can detect mosaicism; and
- can identify structural rearrangements that may be missed by arrays; for example, ring chromosome 20 in epilepsy.
Limitations
Karyotyping:
- is labour intensive;
- has a slow turnaround time;
- is unable to detect uniparental disomy;
- requires dividing cells (and cell culture);
- has much lower resolution than arrays (only changes over 5Mb can be detected);
- carries a risk of cell culture artefacts (abnormal chromosomes arising during cell culture, but not actually present in the patient); and
- carries a (rare) risk that some variants are not detected in cultured cells because they are lost during cell culture (for example, the 12p isochromosome causing Pallister-Killian syndrome is usually lost when blood lymphocytes are cultured).
Practicalities
The samples required for karyotyping depend on the reason for testing.
- Blood samples must be taken into a lithium heparin tube (typically a green-topped tube), which is different to most genomic tests carried out on blood samples. From adults, 5–10ml of blood is required; from children 2–5ml is required; and from babies 1–2ml is required.
- Chorionic villus samples require 12–20mg of cells in a sterile, leak-proof container.
- An amniocentesis sample requires 12–20ml of amniotic fluid in a sterile, leak-proof universal container.
- Products of conception should be in tissue culture media or a dry, sterile container.
- Skin samples should be in tissue culture media or a dry, sterile container.
- Bone marrow aspirate should be collected into a lithium heparin tube (typically a green-topped tube).
The target reporting time from receipt of sample is between 14 and 42 days, depending on the clinical reason for the test.
Key messages
- A karyotype provides a visual, genome-wide screen for large chromosomal variants such as deletions, duplications and structural rearrangements.
- A karyotype is particularly useful for detecting balanced rearrangements that can lead to infertility and structural chromosome rearrangements that result in gene fusions that can drive cancer.
Resources
For clinicians
- Genetic Science Learning Center: Make a karyotype